
Publications
Michael Nash on Google Scholar
#Equally contributing authors; *Corresponding author
Boult, S., Pacak, P., Yang, B., Liu, H., Vogel, V.*, and Nash, M. A.*
Multi-state Catch Bond Formed in the Izumo1:Juno Complex that Initiates Human Fertilization
Nature Communications, 2025, 16: 7952.

Sun, Y., Li, J., Vanella, R., Liu, H., Liu, Z., and Nash, M. A.*
Angewandte Chemie, 2025, e202504483.

Küng, C., Protsenko, O., Vanella, R., and Nash, M. A.*
Protein Science, 2025, 34(5): e70112.

Fernández de Santaella, J., Koch, N. G., Widmer, L., and Nash M. A.*
ACS Chemical Biology, 2025, 20(4): 791-801.

Yang, B., Gomes, D. E. B., Liu, Z., Santos, M. S., Li, J., Bernardi, R. C.* and Nash, M. A.*
ACS Nano 2024, 18, 46, 31912–31922.

Heiniger, M., Vanella,R.*, Walsh-Korb, Z.,* and Nash M. A.
Biomaterials Science & Engineering 2024, 10, 9, 5869–5880

Gomes, D. E. B., Yang, B., Vanella, R., Nash, M. A.*, and Bernardi, R. C.*
Integrating Dynamic Network Analysis with AI for Enhanced Epitope Prediction in PD-L1:Affibody Interactions.
Journal of the American Chemical Society 2024 146, 34, 23842–23853

Liu, Z., Liu, H., Vera, A.M. , Yang, B., Tinnefeld, P.; and Nash, M. A.*
Engineering an artificial catch bond using mechanical anisotropy.
Nature Communications 2024 15, 3019

Vanella, R.*, Küng, C., Schoepfer, A. A., Doffini, V., Ren, J.; and Nash, M. A.*
Understanding activity-stability tradeoffs in biocatalysts by enzyme proximity sequencing.
Nature Communications, 2024, 15: 1807.

Doffini, V., Liu, H., Liu Zh.; and Nash, M. A.*
Iterative Machine Learning for Classification and Discovery of Single-molecule Unfolding Trajectories from Force Spectroscopy Data.
Nano Lett. 2023, 23, 22, 10406–10413

Lopez-Morales, J., Vanella, R., Appelt, E. A., Whillock, S., Paulk, A. M., Shusta, E. V., Hackel, B. J., Liu, C. C.; and Nash, M. A.*
Protein Engineering and High-throughput Screening by Yeast Surface Display: Survey of Current Methods.
Small science, 2023, Vol.3(12)
Küng, Ch.; Vanella, R.; and Nash, M. A.*
Directed evolution of Rhodotorula gracilis D-amino acid oxidase using single-cell hydrogel encapsulation and ultrahigh-throughput screening
React. Chem. Eng. 2023 8, 1960-1968

Liu, H.; Liu, Z.; Santos, M. S.; and Nash, M. A.*
Direct Comparison of Lysine vs. Site-specific Protein Surface Immobilization in Single-molecule Mechanical Assays
Angew. Chem. Int. Ed. 2023 (Accepted)

Lopez-Morales, J.; Vanella, R.; Utzinger, T.; Schittny, V.; Hirsiger, J.; Osthoff, M.; Berger, C.; Guri, Y.; and Nash, M. A.*
Multiplexed on-yeast serological assay for immune escape screening of SARS-CoV-2 variants
iScience 2023 26, 106648.

Fernández De Santaella, J.; Ren, J.; Vanella, R.; Santos, M. S.; and Nash, M. A.*
Enzyme Cascade with Horseradish Peroxidase Readout for High-Throughput Screening and Engineering of Human Arginase-1
Analytical Chemistry 2023, 95 (18), 7150–7157.

Lopez-Morales, J.; Vanella, R.; Kovacevic, G.; Santos, M. S.; and Nash, M. A.*
Titrating Avidity of Yeast-displayed Proteins using a Transcriptional Regulator
ACS Synthetic Biology 202312 (2), 419-431.

Risser, F., Lopez-Morales, J., and Nash, M. A.*
Adhesive Virulence Factors of Staphylococcus aureus Resist Digestion by Coagulation Proteases Thrombin and Plasmin
ACS Bio Med Chem Au, 2022.

Liu, H., Liu, Z., Yang, B., Lopez-Morales, J., and Nash, M. A.*
Optimal Sacrificial Domains in Mechanical Polyproteins: S. epidermidis Adhesins are Tuned for Work Dissipation.
JACS Au, 2(6): 1417-1427, 2022.

Nash, M. A.*
Elastin-like polypeptides: protein-based polymers for biopharmaceutical development
Chimia, 76, 478-479, 2022.
(Editorial: Medicinal Chemistry and Chemical Biology Highlights)

Risser, F., Urosev, I., Lopez-Morales, J., Sun, Y. and Nash, M. A.*
Engineered Molecular Therapeutics Targeting Fibrin and the Coagulation System: a Biophysical Perspective.
Biophysical Reviews, 14, 427-461, 2022.

Santos, M. S., Liu, H., Schittny, V., Vanella, R., and Nash, M. A.*
Correlating Single-molecule Rupture Mechanics with Cell Population Adhesion by Yeast Display
Biophysical Reports, 2(1), 10035, 2022.

Vanella, R., Kovacevic, G. N., Doffini, V., Santaella, J. F., and Nash, M. A.*
High-throughput Screening, Next Generation Sequencing and Machine Learning: Advanced Methods in Enzyme Engineering
Chemical Communications, 58: 2455–2467, 2022.

Nash, M. A.*
Scalable Online Learning in Physical Chemistry
Chimia, 75: 64–66, 2021.

Zhaowei Liu, Rodrigo A. Moreira, Ana Dujmović, Haipei Liu, Byeongseon Yang, Adolfo B. Poma, Michael A. Nash*
Mapping Mechanostable Pulling Geometries of a Therapeutic Anticalin/CTLA-4 Protein Complex
Nano Letters 22(1): 179–187, 2021. bioRxiv 2021.03.09.434559

Huo, Z., Santos, M. S., Drenckhan, A., Holland-Cunz, S., Izbicki, J. R., Nash, M. A., and Gros, S. J.
Metastatic Esophageal Carcinoma Cells Exhibit Reduced Adhesion Strength and Enhanced Thermogenesis
Cells 10(5), 1213; DOI: 10.3390/cells10051213, 2021

Yang, B., Liu, H., Liu, Z., Doenen, R., and Nash, M. A.*
Influence of Fluorination on Single-Molecule Unfolding and Rupture Pathways of a Mechanostable Protein Adhesion Complex
Nano Letters, DOI: 10.1021/acs.nanolett.0c04178; BioRxiv 10.1101/2020.07.09.194894v1, 2020
Associated Data

Urosev, I., Lopez-Morales, J., and Nash, M. A.*
Phase Separation of Intrinsically Disordered Protein-polymers Mechanically Stiffens Fibrin Clots
Advanced Functional Materials, 2020, 2005245; DOI: 10.1002/adfm.202005245

Liu, Z., Liu, H., Vera, A. M., Bernardi, R. C., Tinnefeld, P., and Nash, M. A.*
High Force Catch Bond Mechanism of Bacterial Adhesion in the Human Gut
Nature Communications 11, 4321 (2020), https://doi.org/10.1038/s41467-020-18063-x

Nash, M. A.*
Single-molecule and Single-cell Approaches in Molecular Bioengineering
Chimia, 74:704–709, 2020

Jensen, M. H., Morris, E. J., Tran, H., Nash, M. A., and Tan, C.
Stochastic Ordering of Complexoform Protein Assembly by Genetic Circuits
PLOS Computational Biology, DOI: 10.1371/journal.pcbi.1007997, 2020

Yang, B., Liu, Z., Liu, H., and Nash, M. A.*
Next Generation Methods for Single-molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes
Frontiers in Molecular Biosciences DOI: 10.3389/fmolb.2020.00085, 2020

Nash, M. A.*
Zig Zag AFM Protocol Reveals New Intermediate Folding States of Bacteriorhodopsin
Biophysical Journal, 118(3): 538-540, 2020

Bernardi, R. C.*#, Durner, E.#, Schoeler, C., Malinowska, K. H., Carvalho, B. G., Bayer, E. A., Luthey- Schulten, Z., Gaub, H. E., and Nash, M. A.*
Mechanisms of Nanonewton Mechanostability in a Protein Complex Revealed by Molecular Dynamics Simulations and Single-Molecule Force Spectroscopy
Journal of the American Chemical Society, 141(37): 14752-14763, 2019

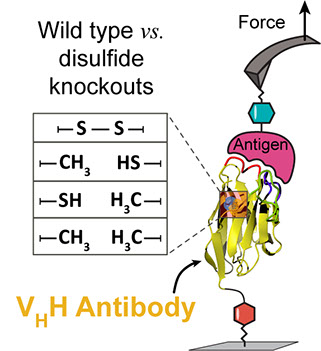
Liu, H., Schittny, V., and Nash, M. A.*
Removal of a Conserved Disulfide Bond Does Not Compromise Mechanical Stability of a VHH Antibody Complex
Nano Letters, 19(8): 5524-5529, 2019
Vanella, R., Ta, D. T., and Nash, M. A.*
Enzyme‐mediated hydrogel encapsulation of single cells for high‐throughput screening and directed evolution of oxidoreductases
Biotechnology and Bioengineering, 116(8): 1878–1886, 2019

Vanella, R., Bazin, A., Ta, D. T., and Nash, M. A.*
Genetically Encoded Stimuli-Responsive Cytoprotective Hydrogel Capsules for Single Cells Provide Novel Genotype−Phenotype Linkage
Chemistry of Materials, 31(6): 1899–1907, 2019

Lewis, D. D., Vanella, R., Vo, C., Rose, L., Nash, M. A., and Tan, C.
Engineered Stochastic Adhesion Between Microbes as a Protection Mechanism Against Environmental Stress
Cellular and Molecular Bioengineering, 11(5): 367-382, 2018

Ta, D. T., Vanella, R., and Nash, M. A.*
Bioorthogonal Elastin-like Polypeptide Scaffolds for Immunoassay Enhancement
ACS Applied Materials & Interfaces, 10(36): 30147-30154, 2018

Gunnoo, M., Cazade, P. A., Orlowski, A., Chwastyk, M., Liu, H., Ta, D. T., Cieplak, M., Nash, M. A., and Thompson, D.
Steered Molecular Dynamics Simulations Reveal the Role of Ca2+ in Regulating Mechanostability of Cellulose-binding Proteins
Physical Chemistry Chemical Physics, 20(35): 22674-22680, 2018

Liu, H.#, Ta, D. T.#, and Nash, M. A.*
Mechanical Polyprotein Assembly using Sfp and Sortase-mediated Domain Oligomerization for Single-molecule Studies
Small Methods, 2(6): 1800039, 2018

Ta, D. T., Vanella, R., and Nash, M. A.*
Magnetic Separation of Elastin-like Polypeptide Receptors for Enrichment of Cellular and Molecular Targets
Nano Letters, 17(12): 7932-7939, 2017

Verdorfer, T., Bernardi, R., Meinhold, A., Ott, W., Luthey-Schulten, Z., Nash, M. A.*, and Gaub. H. E.
Combining in Vitro and in Silico Single Molecule Force Spectroscopy to Characterize and Tune Cellulosomal Scaffoldin Mechanics
Journal of the American Chemical Society, 139(39): 17841-17852, 2017

Ott, W.#, Jobst, M. A.#, Bauer, M., Durner, E., Milles, L., Nash, M. A., and Gaub. H. E.
Elastin-like Polypeptide Linkers for Single Molecule Force Spectroscopy
ACS Nano, 11 (6): 6346-6354, 2017
Durner, E., Ott, W., Nash, M. A., and Gaub, H. E.
Post-translational Sortase-mediated Attachment of High-strength Force Spectroscopy Handles
ACS Omega, 2(6): 3064-3069, 2017

Ott, W., Jobst, M. A., Schoeler, C., Gaub, H. E., and Nash, M. A.*
Single-Molecule Force Spectroscopy on Polyproteins and Receptor-Ligand Complexes: The Current Toolbox
Journal of Structural Biology, 197(1): 3-12, 2017

Milles, L., Bayer, E. A., Nash, M. A., and Gaub, H. E.
Mechanical Stability of a High-Affinity Toxin Anchor From the Pathogen Clostridium Perfringens
Journal of Physical Chemistry B, 121(15): 3620-3626, 2017

Nash, M. A.*
Interfacial Polymerization Systems for Screening of Multi-enzyme Complexes
European Cells and Materials, 32(Supp. 5): 33, 2016
Schoeler, C., Verdorfer, T., Gaub, H. E., and Nash, M. A.*
Biasing Effects of Receptor Ligand Complexes on Protein Unfolding Statistics
Physical Review E, 94: 042412, 2016
Nash, M. A.*, Smith, S. P., Fontes, M. G. A. C., and Bayer, E. A.
Single- versus Dual-binding Conformations in Cellulosomal Cohesin-dockerin Complexes
Current Opinion in Structural Biology, 40:89-96, 2016
Ott, W., Nikolaus, T., Gaub, H. E., and Nash, M. A.*
Sequence Independent Cloning and Post-translational Modification of Repetitive Protein Polymers through Sortase and Sfp-mediated Enzymatic Ligation
Biomacromolecules, 17(4): 1330-1338, 2016
Nash, M. A.* and Shoseyov, O.
Editorial: Nanobiotechnology at a Crossroads: Moving Beyond Proof-of-Concept
Current Opinion in Biotechnology, 39:1-3, 2016
Malinowska, K. H., and Nash, M.A.*
Enzyme- and Affinity Biomolecule-mediated Polymerization Systems for Biological Signal Amplification and Cell Screening
Current Opinion in Biotechnology, 39:68-75, 2016
Gunnoo, M., Cazade, P. A., Galera-Prat, A., Nash, M. A., Czjzek, M., Cieplak, M., Alvarez, B., Aguilar, M., Karpol, A., Gaub, H. E., Carrion-Vazquez, M., Bayer, E. A., and Thompson, D.
Nanoscale Engineering of Designer Cellulosomes
Advanced Materials, 28: 5619-5647, 2016
Jobst, M., Milles, L., Schoeler, C., Ott, W., Fried, D. B., Bayer, E. A., Gaub, H. E., and Nash, M. A.*
Resolving Dual Binding Conformations of Cellulosome Cohesin-Dockerin Complexes using Single-Molecule Force Spectroscopy
eLife, DOI: 10.7554/eLife.10319, 2015
Schoeler, C., Bernardi, R. C., Malinowska, K. M., Durner, E., Ott, W., Bayer, E. A., Schulten, K., Nash, M. A.*, and Gaub, H. E.
Mapping Mechanical Force Propagation Through Biomolecular Complexes
Nano Letters, 15(11): 7370-7376, 2015
[Featured on journal cover; Highlighted on nanotechweb.org & physicsworld.com]
Malinowska, K.H., Rind, T., Verdorfer, T., Gaub, H.E., and Nash, M. A.*
Quantifying Synergy, Thermostability, and Targeting of Cellulolytic Enzymes and Cellulosomes with Polymerization-Based Amplification
Analytical Chemistry, 87(14):7133-7140, 2015
Nash, M. A.*, Bernardi, R., Schoeler, C., Malinowska, K. H., Gaub, H., Schulten, K., and Bayer, E.
Analyzing Force Propagation Through a Stable Protein Complex with Experiments and Simulation
European Biophysical Journal, 44 (Suppl 1): S43–S248, O-307, 2015
Schoeler, C.# Malinowska, K. H.,# Bernardi, R. C., Milles, L. F., Jobst, M. A., Durner, E., Ott, W., Fried, D. B., Bayer, E. A., Schulten, K., Gaub, H. E., and Nash, M. A.*
Ultrastable Cellulosome-Adhesion Complex Tightens Under Load
Nature Communications, 5: 1-35, 2014
Otten, M.,# Ott, W.,# Jobst, M. A.,# Milles, L., Verdorfer, T., Pippig, D., Nash, M. A.*, and Gaub, H.E.
From Genes to Protein Mechanics on a Chip
Nature Methods, 11(11): 1127-1130, 2014
[Highlighted in Nat. Meth. News & Views (subscription required); Cell Press Chemistry & Biology Select]
Malinowska, K., Verdorfer, T., Meinhold, A., Milles, L.F., Funk, V., Gaub, H.E., and Nash, M. A.*
Redox-Initiated Hydrogel System for Detection and Real-Time Imaging of Cellulolytic Enzyme Activity
ChemSusChem, 7(10): 2825-2831, 2014
[Featured on journal cover]
Wolff, M., Braun, D., and Nash, M. A.*
Detection of Thermoresponsive Polymer Phase Transition in Dilute Low-Volume Format by Microscale Thermophoretic Depletion
Analytical Chemistry, 86(14): 6797–6803, 2014
[Featured on journal cover]
Jobst, M. A., Schoeler, C., Malinowska, K., and Nash, M. A.*
Investigating Receptor-Ligand Systems of the Cellulosome using AFM-based Single-molecule Force Spectroscopy
Journal of Visualized Experiments, 82, e50950, 2013
Nash, M. A.*, and Gaub, H. E.
Single-Molecule Adhesion of a Stimuli-Responsive Oligo(ethylene glycol) Co-polymer to Gold
ACS Nano, 6(12): 10735–10742, 2012
[Featured on journal cover]
Stahl, S. W.#, Nash, M. A.#,*, Fried, D. B, Slutzki, M., Barak, Y., Bayer, E. A., and Gaub, H. E.
Single-Molecule Dissection of the High-Affinity Cohesin-Dockerin Complex
PNAS, 109(50): 20431–20436, 2012
Nash, M. A., Waitumbi, J., Yager, P., Hoffman, A. S., and Stayton, P. S.
Multiplexed Enrichment and Detection of Malarial Biomarkers using a Stimuli-Responsive Iron Oxide and Gold Nanoparticle Reagent System
ACS Nano, 6(8): 6776–6785, 2012
Nash, M. A., Golden, A., Hoffman, J. M., Lai, J. J., and Stayton, P. S.
Smart Surfaces for Point-of-Care Diagnostics
Intelligent Surfaces in Biotechnology. John Wiley & Sons. Grandin, H. M., Textor, M. (Editors) & and Whitesides, G. M. (Foreword), 2012
[Featured on book cover]
Nash, M. A.
Mixed Nanoparticle System for Purification, Enrichment, and Detection of Biomarkers
University of Washington, Seattle – 2011 PhD Dissertation
Nash, M. A.*
Thermal Depolymerization
Green Technology: Green Series Volume 10, 2011 SAGE Publications. Mulvaney, D.R. (Editor)
Nash, M. A.*
Biomimetics
Encyclopedia of Nanoscience and Society, 2010 SAGE Publications. Guston, D.H. (Editor)
Nash, M. A., Yager, P., Hoffman, A. S., and Stayton, P. S.
Magnetic Enrichment of “Smart” Gold Nanoparticle-Antibody Conjugates for High-Sensitivity Diagnostic Immunochromatography
European Cells and Materials, 20(SUPPL. 3): 184, 2010
Nash, M. A., Lai, J. J., Hoffman, A. S., Yager, P., and Stayton, P. S.
“Smart” Diblock Copolymers as Templates for Magnetic-Core Gold-Shell Nanoparticle Synthesis
Nano Letters, 10(1): 85–91, 2010
Nash, M. A., Yager, P., Hoffman, A. S., and Stayton, P. S.
Mixed Stimuli-Responsive Magnetic and Gold Nanoparticle System for Rapid Purification, Enrichment, and Detection of Biomarkers
Bioconjugate Chemistry, 21(12): 2197–2204, 2010
Nash, M. A.*, Hoffman, J. M., Stevens, D., Hoffman, A. S., Stayton, P. S., and Yager, P.
Laboratory Scale Protein Striping System for Patterning Biomolecules onto Paper Based Immunochromatographic Test Strips
Lab on a Chip, 10(17): 2279–2282, 2010
Warner, C. L., Addleman, R. S., Cinson, A. D., Droubay, T. C., Engelhard, M. H., Nash, M. A., Yantasee, W., and Warner, M. G.
High Performance Superparamagnetic Nanoparticle Based Heavy Metal Sorbents for Removal of Contaminants from Natural Waters
ChemSusChem, 3(6): 749–757, 2010
[Featured on journal cover]
Lai, J. J., Nelson, K. E., Nash, M. A., Hoffman, A. S., Yager, P., and Stayton, P. S.
Dynamic Bioprocessing and Microfluidic Transport Control with Smart Magnetic Nanoparticles in Laminar-Flow Devices
Lab on a Chip, 9(14): 1997–2002, 2009
Malmstadt, N., Nash, M. A., Purnell, R. F., and Schmidt, J. J.
Automated Formation of Lipid-Bilayer Membranes in a Microfluidic Device
Nano Letters, 6(9): 1961–1965, 2006

